Coiled coils as a model for protein folding
The molecular basis of a protein is a single polypeptide chain made of amino acids. The sequence of the amino acids determines how the protein folds into its three dimensional structure but the underlying mechanisms are very complex. Coiled coils are a good model system to study protein folding, since they are structurally very simple. They consist of two alpha helices that wrap around each other, held together via a hydrophobic core of amino acids in the contact zone (red circles named a and d)
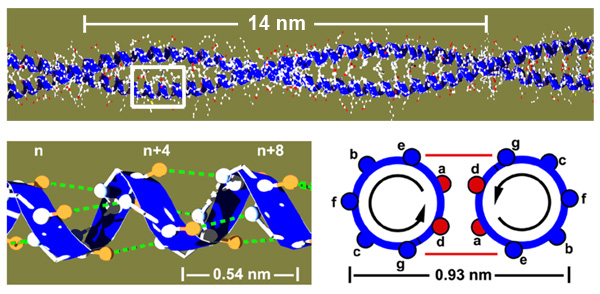
Coiled coil mechanics in thermodynamic equilibrium
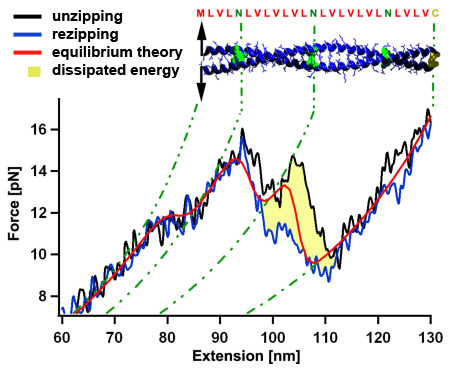
Mechanical unzipping of a model coiled coil with almost perfect hydrophobic core shows a force plateau with force fluctuations around 12 pN. Because the unzipping and rezipping traces show only a very small hysteresis of a few kT (yellow area) we can use a thermodynamic equilibrium description (red) showing that the force fluctuations are related to distinct hydrophilic amino acids (N) within the coiled coil core. When the coiled coil first folds, it has to built a nucleation seed consisting of four alpha helices, which leads to the more pronounced force dip where energy is dissipated to the heat bath. Since all atomic interactions of the coiled coil are known, we can understand the energetic contributions leading to its three dimensional fold in high detail. [read more]
From equilibrium towards non-equilibirum
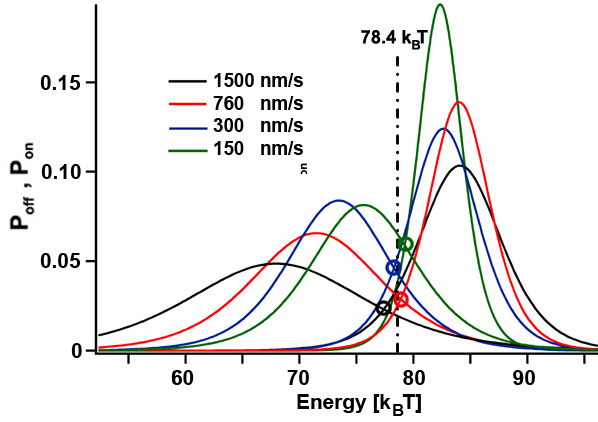
The folding of coiled coils is not completely in thermodynamic equilibrium since a few kT are dissipated to the heat bath per unzipping/rezipping cycle (yellow area). If the pulling speed of the unzipping/rezipping cycles is further increased, more energy gets dissipated and the system gets pushed further towards non-equilibrium. Since we can determine the energy needed to unfold the structure and the energy gained during folding of the coiled coil (histograms for Poff and Pon) for different cycle speeds, we can experimentally test fluctuation theorems that link the non-equilibirum regime with equilibrium thermodynamics. [read more]
Designing the mechanics of coiled coils
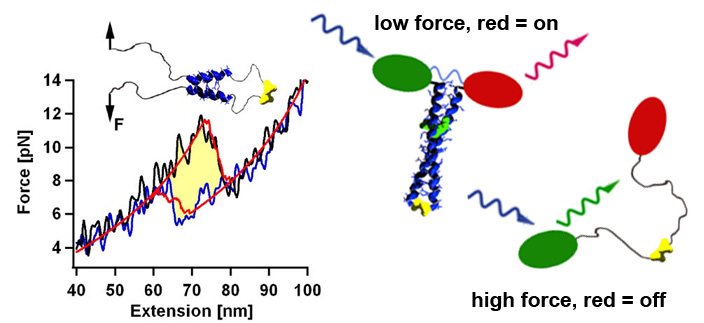
Using the thermodynamic model that relates the amino acid sequence to the mechanical stability and folding dynamics, we can design now coiled coils with a desired stability profile. For example, by adjusting the size of the nucleation seed we can tune the refolding forces in a range between 2 and 8 pN. Thus we could construct in vivo biosensors that could report cell internal forces via a change in the emission wavelength. [read more]